RootsWeb:
GENEALOGY-DNA-L [DNA] Y-STR database issues - [ このページを訳す BETA
]
... They listed. DYS 389i =
10 DYS 389ii-i = 16 Since DYS 389ii-i = 16, that means that
16 is the difference between the value of DYS389ii and DYS389i.
Since DYS389i =
10, then you must add this value to the value of DYS389ii-i to obtain
the value ...
archiver.rootsweb.com/th/
read/GENEALOGY-DNA/2001-07/0996535491 - 6k - 補足結果 -
RootsWeb: MEXICO-L [MEX] Re: TRUJILLO
FAMILY -
[ このページを訳す BETA
]
... Mutations at positions
(129, 256, 270, 399) Oxford Ancestors Y-STR signature:
DYS394/19 15 DYS388 12 DYS390 24 DYS391 11 DYS392 14 DYS393 13 DYS389i 10 DYS389ii
16 DYS425 12 DYS426 12 Relative Genetics/Ancestry.com paternal
signature: DYS385a ...
archiver.rootsweb.com/th/read/MEXICO/2002-08/1029882222
- 17k - 補足結果 -
[ 他、archiver.rootsweb.com内のページ
]
www.OxfordAncestors.com ::
View topic - 389ii - [ このページを訳す BETA
]
... So, for example, if you
have ten (10) at DYS389i and sixteen (16) DYS389ii, then
you would input twenty-nine (29) for DYS389ii; the sum of 10 + 16
+ 3. The reason
for this modification is that Oxford Ancestors reports the actual value for the
...
www.oxan.click2.co.uk/cgi-bin/viewtopic.
php?p=2829&sid=01303d44ece7a9cd48deb4e1ac2ebe20 - 27k - 補足結果 -
[PDF] Basic Principles of Forensic
Basic Principles of Forensic ...
ファイルタイプ: PDF/Adobe Acrobat
... genetic information from one homologous chromosome to another (ie no
crossing over)
Page 15. Page 16. evolution of the NRY sequence classes Page 17. Page
18. Page 19. ...
DYS392 DYS392 DYS391 DYS391 DYS390 DYS390 DYS19 DYS19 DYS389I DYS389I DYS389II
...
www.promega.com/geneticidproc/
ussymp15proc/workshops/y-chromosomestrs.pdf - 補足結果 -
Skadi Forum Archive - Y
chromosome STRs in Croatians. - [ このページを訳す BETA
]
... The most frequent DYS19
allele was 16, while at the DYS389II the most frequent were
alleles 30 and 31. The most frequent Y chromosome haplotype (16-13-13-31-24-11-11-
13) was found in 33 individuals (7.22%). One hundred and seventy-four haplotypes
...
forum.skadi.net/archive/topic/14096-1.html
- 4k - 補足結果 -
DNA Heritage - [ このページを訳す BETA
]
The other
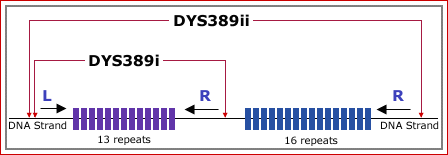
segment, DYS389ii, is much longer and brackets both the 13 repeats and
the 16 repeat sections. ... DYS389ii = 30 (14 + 16)
Both repeat numbers have gone
up, but only one mutation has actually occurred. ...
www.dnaheritage.com/markers.asp
- 25k -
DYS389i and DYS389ii
Combinations in the European Y-STR Spreadsheet - [ このページを訳す BETA
]
Number of
occurrence of each combination of DYS389i and DYS389ii in the European
Y-STR Database http://www.ystr.org/europe Database queried 1/10/2002. Raw Data.
raw data graph ... 16, 0, 0, 0, 0, 0, 0, 0.000233, 0, 0, 0.000233
...
www.contexo.info/FrequencyCharts/E389Combined.htm
- 108k -
[PDF] Amplification of Y-chromosomal
STRs from ancient skeletal material
ファイルタイプ: PDF/Adobe Acrobat
... (–) indicates that no result could be obtained owing to
amplification failure of
the corresponding STR system. Sample. Extract. DYS389I. DYS19. DYS390. DYS389II.
(145–169 bp). (174–210 bp). (191–227 bp). (259–291 bp). Do 1076. A. 12. 16.
25 ...
www.springerlink.com/index/862T8NN34NRCB3NA.pdf
- 補足結果 -
What if there is a 10/12 match - [ このページを訳す BETA
]
... However, there can be a
mutation at DYS389II that will not affect DYS389I]. As for
Duerinckx, we have had a further
test done. ... Out
of 30 Germans (Bavaria was another
category), 40% were Hg 1, 20% Hg
2, 30% Hg 3, 3% Hg 9, 3% Hg 16, and 3% Hg ...
home.earthlink.net/~wwhiteside/ WhitesideDNA/Whitesidepg3.htm - 16k - 補足結果 -
Ybase :: genealogy by numbers - [ このページを訳す BETA ]
DYS389i. DYS389ii. DYS390. DYS391. DYS392.
DYS393. DYS426. DYS437. etc... 001.
Search Ybase. 14. 15. 18. 16. 15. 30. 23. 10. 12. 13. 12. 15 . 002. Search
Ybase
... 16 . 008. Search Ybase. 12. 15. 15. 17. 12. 29. 22. 12. 10.
13. 12. 16 . ...
www.ybase.org/haplomatic.asp
- 37k - 2006年2月11日 -
|
|
|
|
||||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
43 Y-chromosome markers make up the Y-DNA test. They have |
|
|
|||||||||||||||||||||
|
|
all been selected for their
variability between and within different populations around the world, as
recognised in several recent scientific publications. |
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
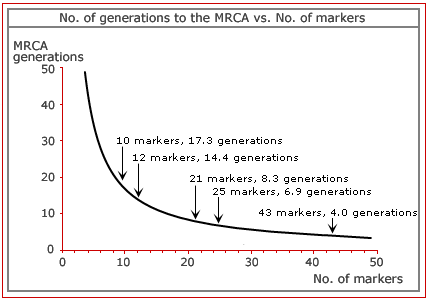
So, how many markers make a good test? |
|
|
|||||||||||||||||||||
|
|
Generally, having more markers to a
test is best. Too few test markers, e.g. 10 or 12,
can often give inconclusive results. These 'low-resolution' tests can really
only confirm that two people are NOT related, thus the vast majority of genealogists
find they need to upgrade to a more accurate test anyway. Because the
low-resolution tests are of little value to genealogists, we won't offer
them. High-resolution tests, i.e. 20+, are enough to answer your genealogical
questions. Our promotional test of 43 markers is a vast increase on that
figure, and also allows you to compare yourself with newer databases that
have come online as well as any future databases. With 43 markers, there
shouldn't ever be a need to upgrade. |
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
Each
individual marker is pinpointed using 'primer-pairs'. These primer- |
|
|
|||||||||||||||||||||
|
|
pairs are custom-made chemicals which
attach themselves to either side of the marker to be studied. Adding enzymes
and several additional ingredients, the marker is 'amplified'. |
|
|
|||||||||||||||||||||
|
|
|
|
||||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
Multiplex
reactions allow several markers to be amplified at the same |
|
|
|||||||||||||||||||||
|
|
time, drastically reducing the time
and effort needed to test for the total of 43 markers. |
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
The markers
used by DNA Heritage are: |
|
|
|||||||||||||||||||||
|
|
http://wanclik.free.fr/tree_r1a.htm |
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
43 markers
is enough markers to be in a position to best answer your |
|
|
|||||||||||||||||||||
|
|
genealogical questions. The graph below
comparing the time to the MRCA vs. number of markers essentially shows the
accuracy of the tests that are currently available. Basically, the less time
to the MRCA, the better the test. |
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|||||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
DYS389 interpretation: |
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|||||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
One of the primer-pairs for DYS389, Primer L, has a unique binding site. However,
the other primer, Primer R, binds at two sites. The other segment, DYS389ii, is much
longer and brackets both the 13 repeats and the 16 repeat sections. Given the above scenario, Tw DYS 389i 13 Tw Dys 389ii 16 |
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
|
|
|
|||||||||||||||||||||
|
|
The results you receive from DNA Heritage will come with full |
|
|
|||||||||||||||||||||
|
|
interpretation. |
|
|
|||||||||||||||||||||
|
Number of occurrence of each
combination of DYS389i and DYS389ii Raw Data |
|||||||||||||||||||||||||
|
DYS389ii
Most frequent |
|||||||||||||||||||||||||
|
DYS389i |
26 |
27 |
28 |
29 |
30 |
31 |
32 |
33 |
34 |
Totals |
|||||||||||||||
|
9 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
0 |
0 |
1 |
|||||||||||||||
|
10 |
3 |
0 |
0 |
0 |
1 |
0 |
0 |
0 |
0 |
4 |
|||||||||||||||
|
11 |
1 |
12 |
11 |
1 |
0 |
0 |
0 |
0 |
0 |
25 |
|||||||||||||||
|
12 |
2 |
50 |
965 |
494 |
154 |
29 |
5 |
0 |
0 |
1699 |
|||||||||||||||
|
13 |
5 |
17 |
345 |
2823 |
1607 |
501 |
95 |
10 |
0 |
5403 |
|||||||||||||||
|
14 |
0 |
3 |
15 |
110 |
761 |
370 |
128 |
14 |
0 |
1401 |
|||||||||||||||
|
15 |
0 |
0 |
1 |
1 |
4 |
30 |
17 |
1 |
1 |
55 |
|||||||||||||||
|
16 |
0 |
0 |
0 |
0 |
0 |
0 |
2 |
0 |
0 |
2 |
|||||||||||||||
|
17 |
0 |
0 |
0 |
0 |
1 |
0 |
0 |
1 |
0 |
2 |
|||||||||||||||
|
Totals |
11 |
82 |
1338 |
3429 |
2528 |
930 |
247 |
26 |
1 |
8592 |
|||||||||||||||
|
|
|||||||||||||||||||||||||
|
|
|||||||||||||||||||||||||
|
DYS389i |
26 |
27 |
28 |
29 |
30 |
31 |
32 |
33 |
34 |
Totals
|
|||||||||||||||
|
9 |
0.000000 |
0.000000 |
0.000116 |
0.000000 |
0.000000 |
0.000000 |
0.000000 |
0.000000 |
0.000000 |
0.000116 |
|||||||||||||||
|
10 |
0.000349 |
0.000000 |
0.000000 |
0.000000 |
0.000116 |
0.000000 |
0.000000 |
0.000000 |
0.000000 |
0.000466 |
|||||||||||||||
|
11 |
0.000116 |
0.001397 |
0.00128 |
0.000116 |
0 |
0 |
0 |
0 |
0 |
0.002910 |
|||||||||||||||
|
12 |
0.000233 |
0.005819 |
0.112314 |
0.057495 |
0.017924 |
0.003375 |
0.000582 |
0 |
0 |
0.197742 |
|||||||||||||||
|
13 |
0.000582 |
0.001979 |
0.040154 |
0.328561 |
0.187034 |
0.05831 |
0.011057 |
0.001164 |
0 |
0.628841 |
|||||||||||||||
|
14 |
0 |
0.000349 |
0.001746 |
0.012803 |
0.088571 |
0.043063 |
0.014898 |
0.001629 |
0 |
0.163059 |
|||||||||||||||
|
15 |
0 |
0 |
0.000116 |
0.000116 |
0.000466 |
0.003492 |
0.001979 |
0.000116 |
0.000116 |
0.006401 |
|||||||||||||||
|
16 |
0 |
0 |
0 |
0 |
0 |
0 |
0.000233 |
0 |
0 |
0.000233 |
|||||||||||||||
|
17 |
0 |
0 |
0 |
0 |
0 |
0 |
0.000233 |
0 |
0 |
0.000233 |
|||||||||||||||
|
Totals |
0.001280 |
0.009544 |
0.155726 |
0.399092 |
0.294227 |
0.108240 |
0.028748 |
0.003026 |
0.000166 |
1.000000 |
|||||||||||||||
|
|
|
|||||||||||||||||||||||
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
|